I had to make a presentation for one of my chemistry classes here, and they told us to use the excellent (but Win32/Mac-only) ChemDraw application for making our own chemical structures. Since I don’t own a Mac, that didn’t work out, so I went looking for alternatives.
While searching, I found that it is not that they don’t exist. Quite the contrary. It’s just nearly impossible to find what you’re looking for! My first step was to look on the Bioinformatics-for-Linux homepage and see if anything exists over there (I found this page after Googling for ‘linux molecule structures’ or so). The best I could find on that page was RasMol, which is some 60s hippie-style half-console application with its own scripting interface (i.e. you don’t draw atoms; you give it a command to draw an atom or a bond). Eh, whazeggie? Right. After reading the documentation for half an hour, I still didn’t know how to draw the first C atom on the drawingboard and decided to give up and look elsewhere; so I googled for ‘chemdraw linux’, and found an excellent review comparing several of the alternatives. After spending nearly half an hour first trying to compile XDrawChem (which appeared to require Qt, so I stopped), I found a winner: GChemPaint! Lovely integrated in my desktop (but why the toolbar/statusbar?), took me 10 minutes to make RPMs for some dependencies, compiled and simply just works.
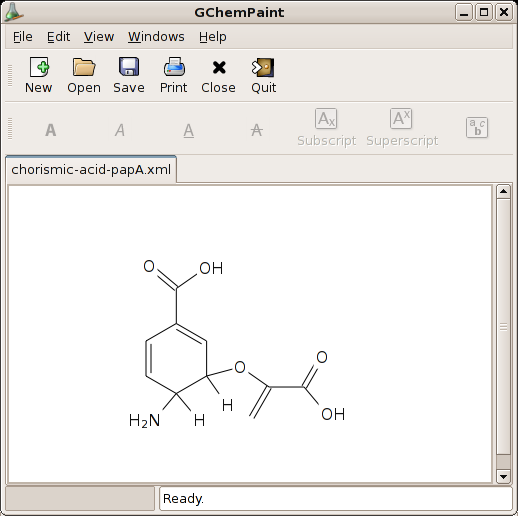
Although it doesn’t do everything I need, it does most of it, and the missing bits can be added by hand using some general-purpose drawing application like the GIMP later on.
